 1.1 Search Functions
1.1 Search Functions
TPIA2 provides seven methods to search the functions of Camellia genes of interest.
1.1.1 Locus search
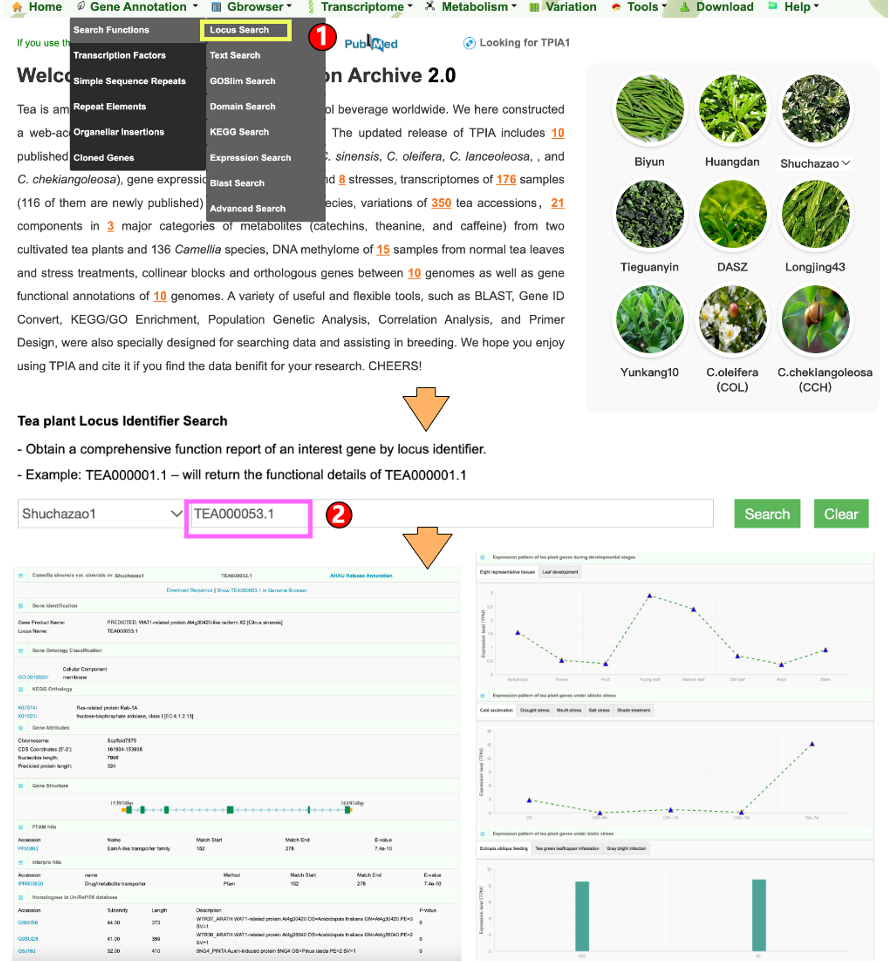
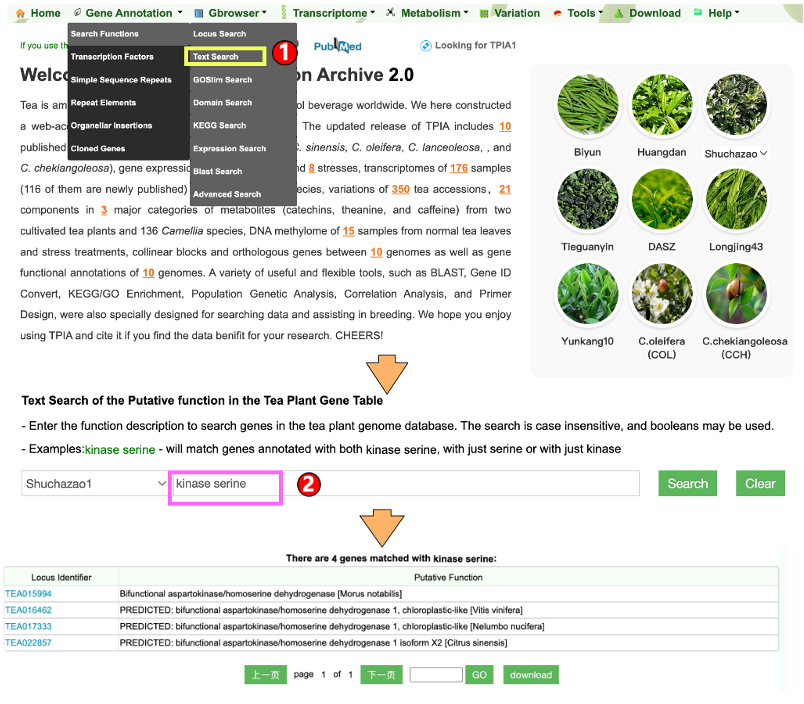
1.1.2 Text search
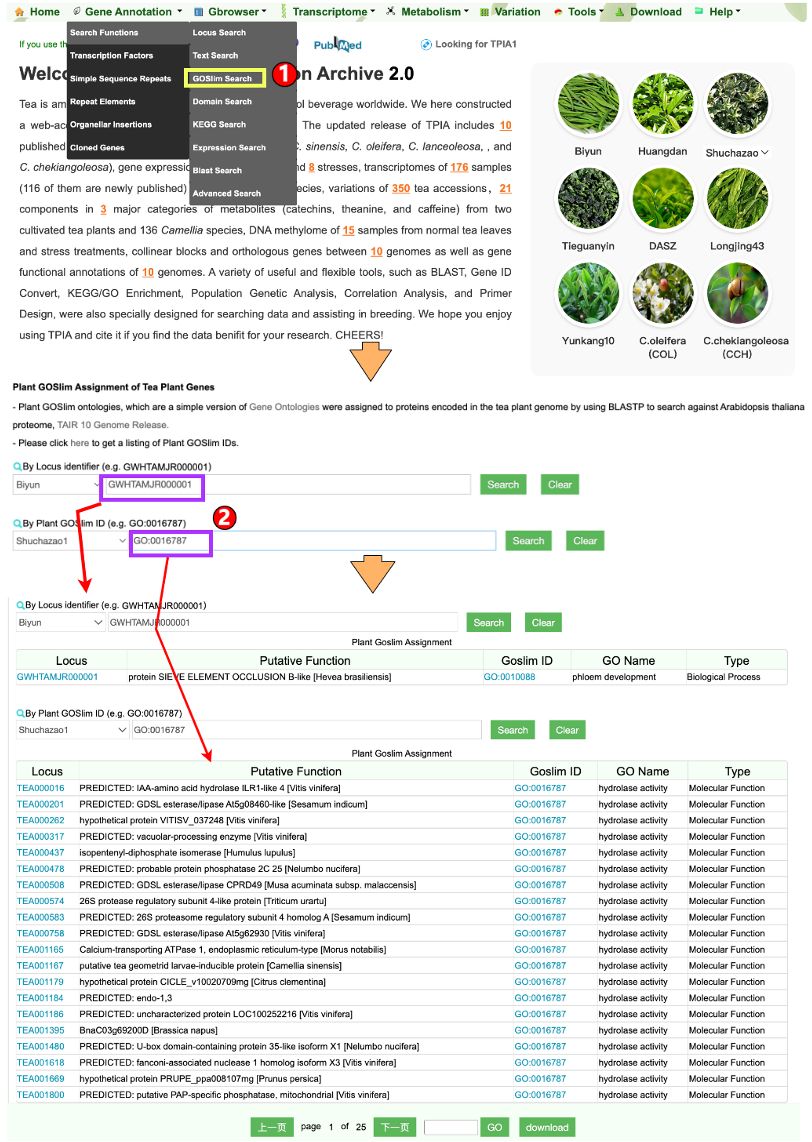
1.1.3 GOSlim Search
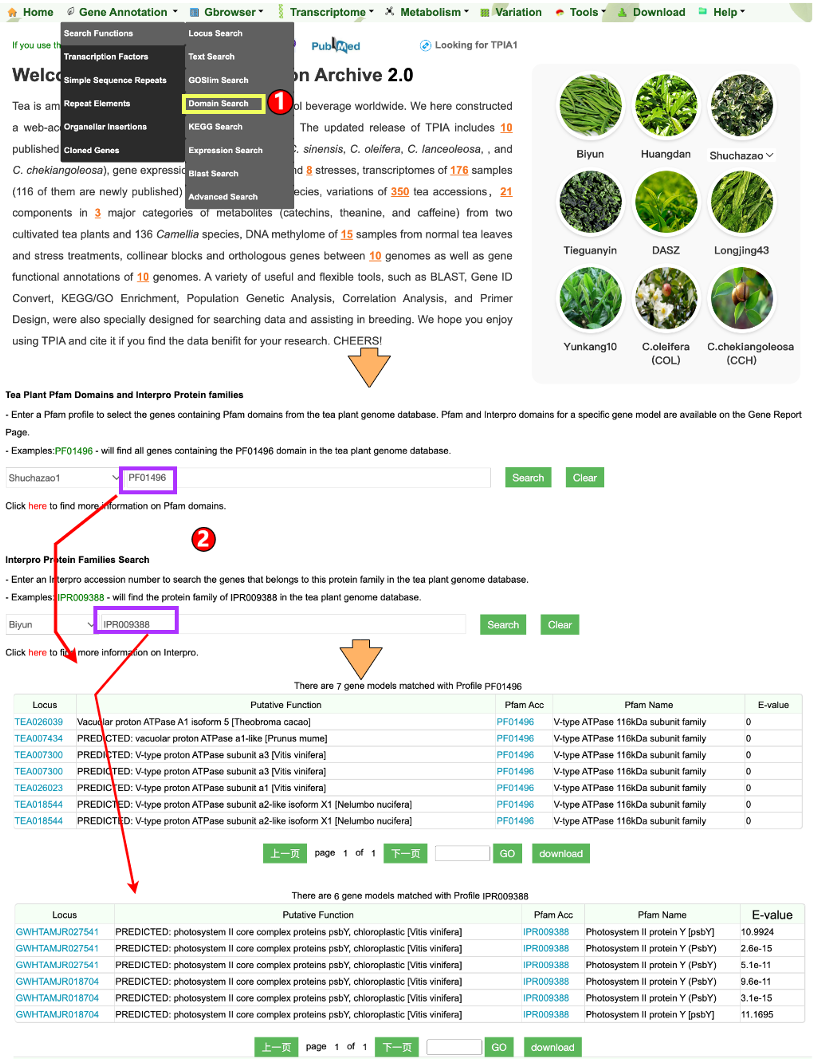
1.1.4 Domain Search
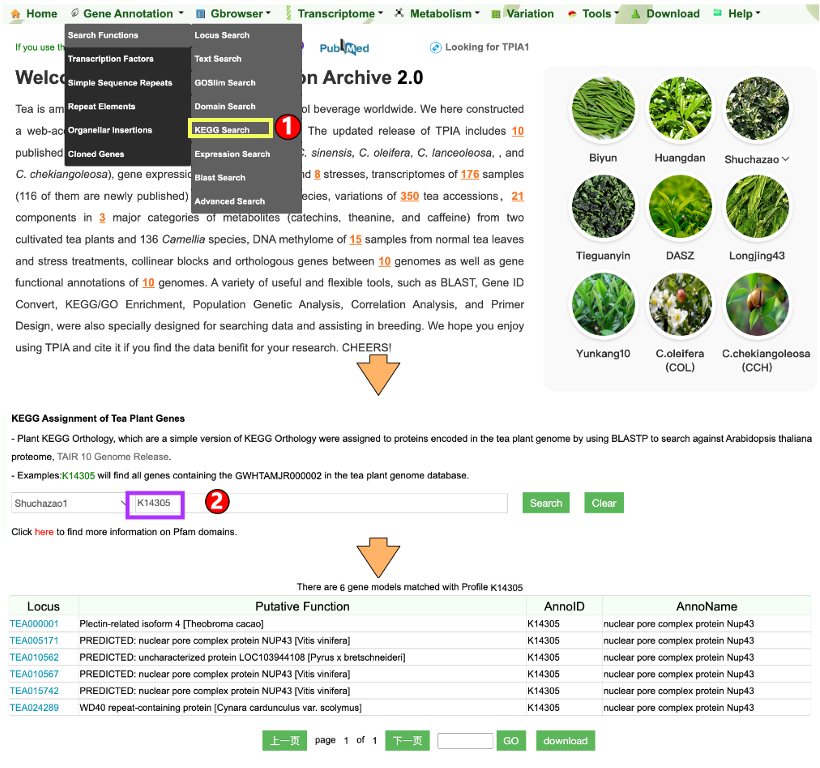
1.1.5 KEGG Search
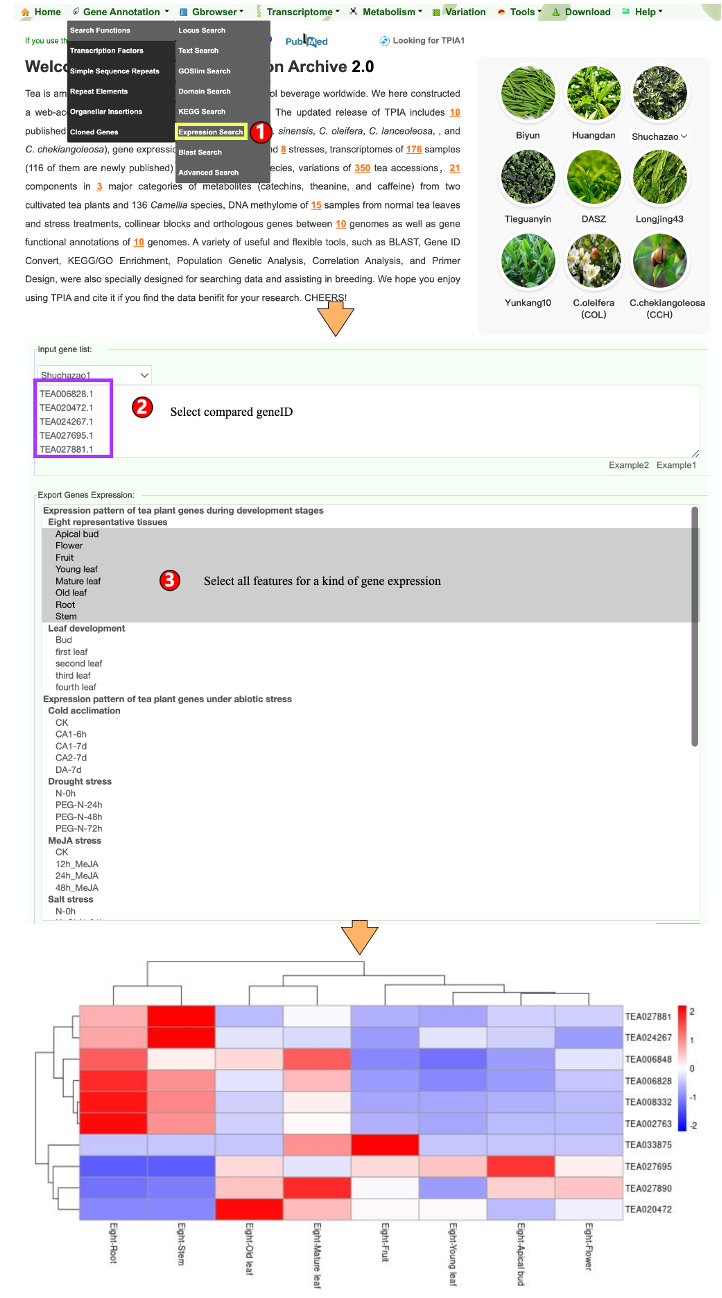
1.1.6 Expression Search
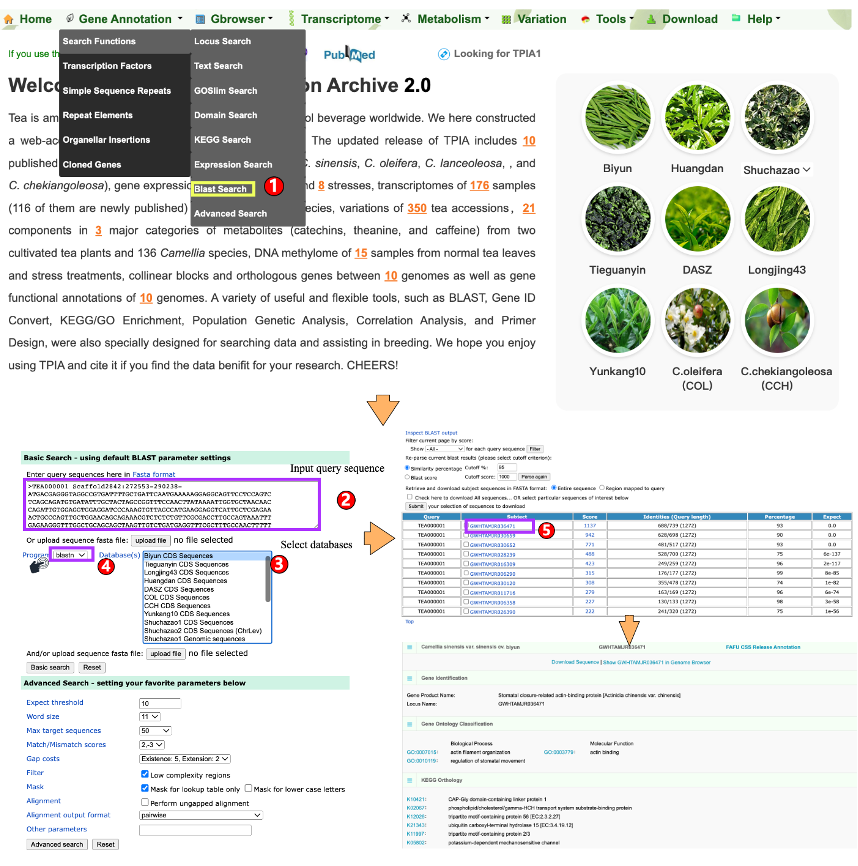
1.1.7 Blast Search
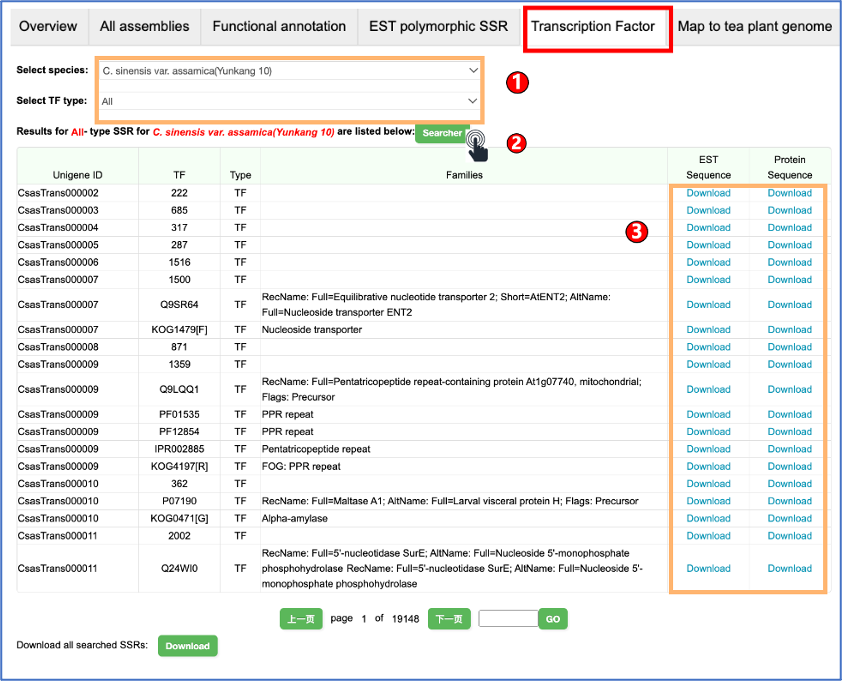
 1.2 Transcription Factor
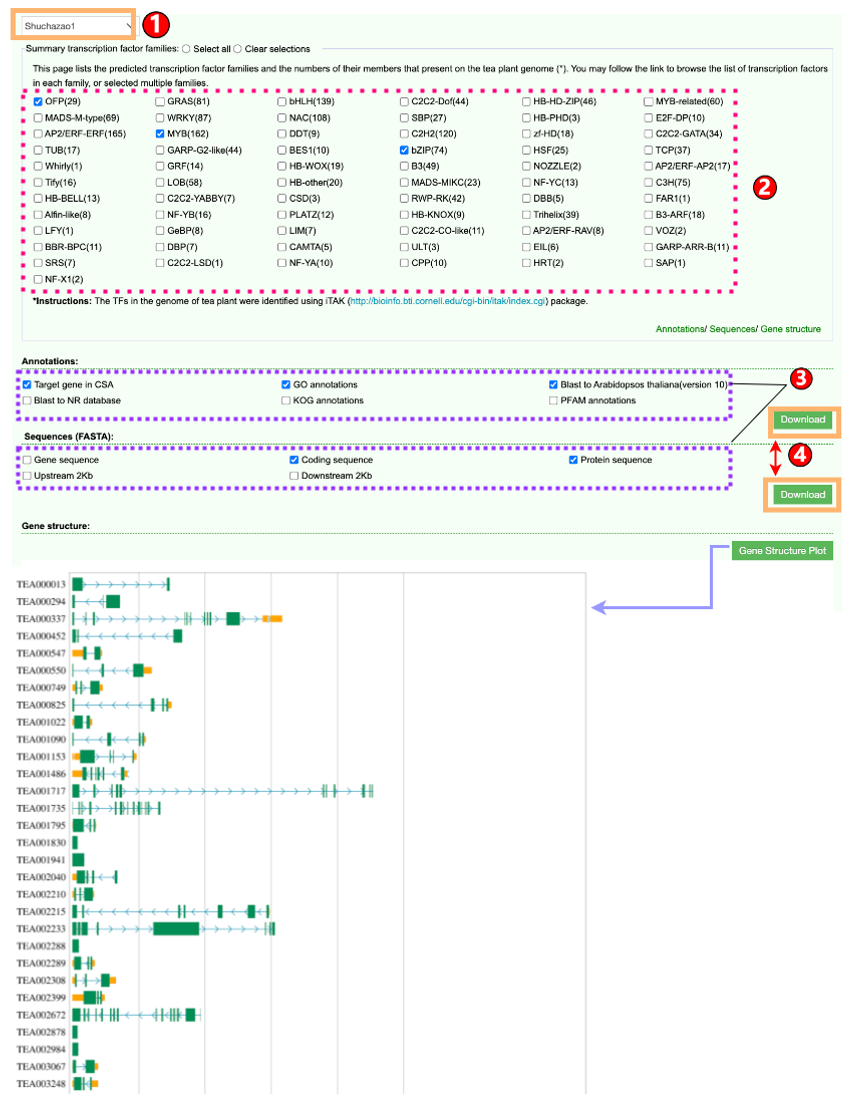
1.2 Transcription Factor
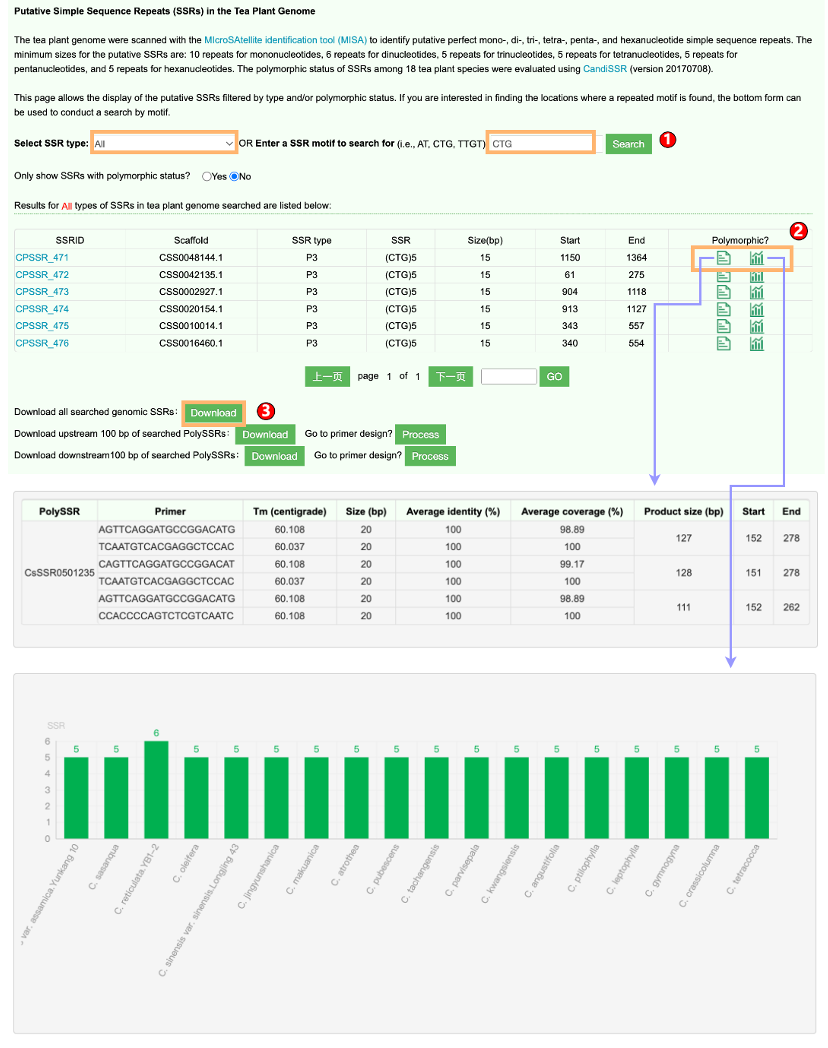
 1.3 Simple Sequence Repeats
1.3 Simple Sequence Repeats
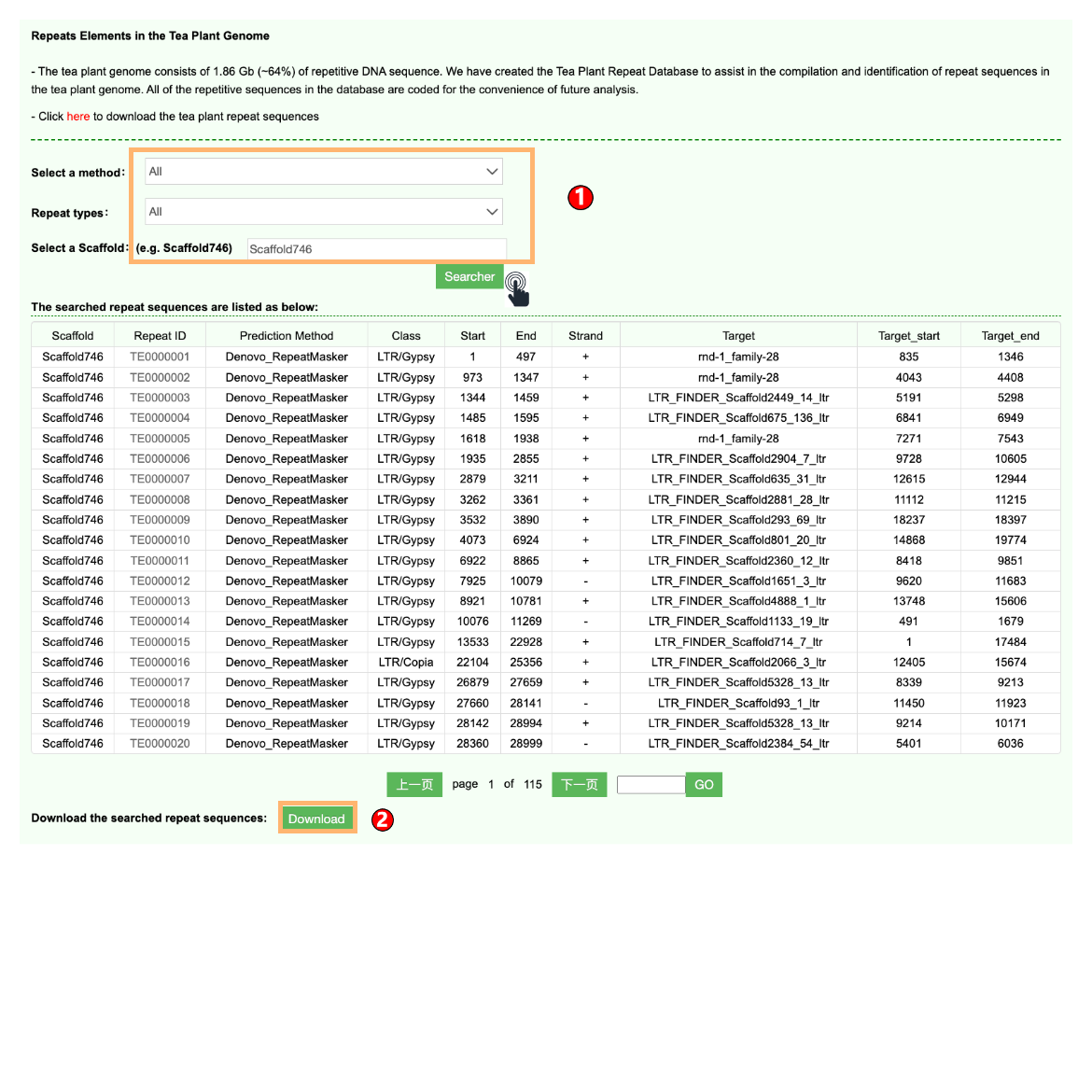
 1.4 Repeats Elements
1.4 Repeats Elements
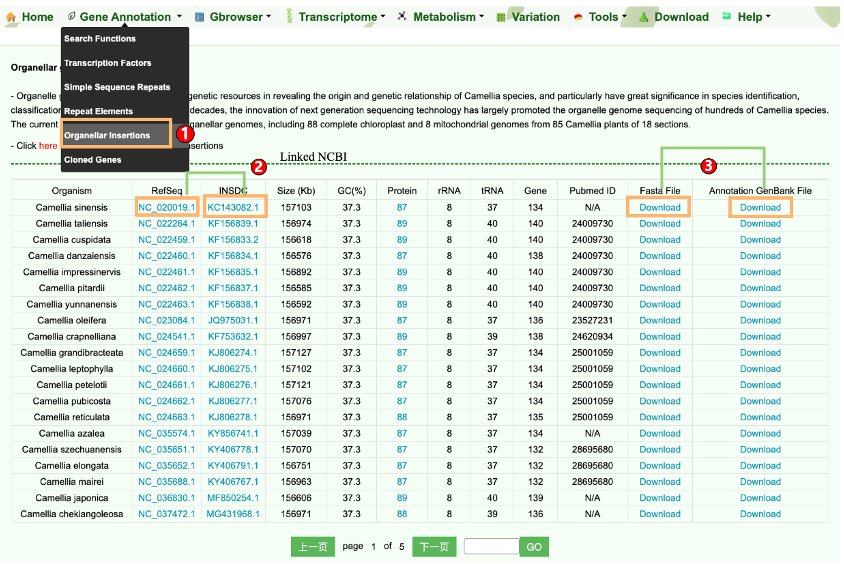
 1.5 Organellar Insertions
1.5 Organellar Insertions
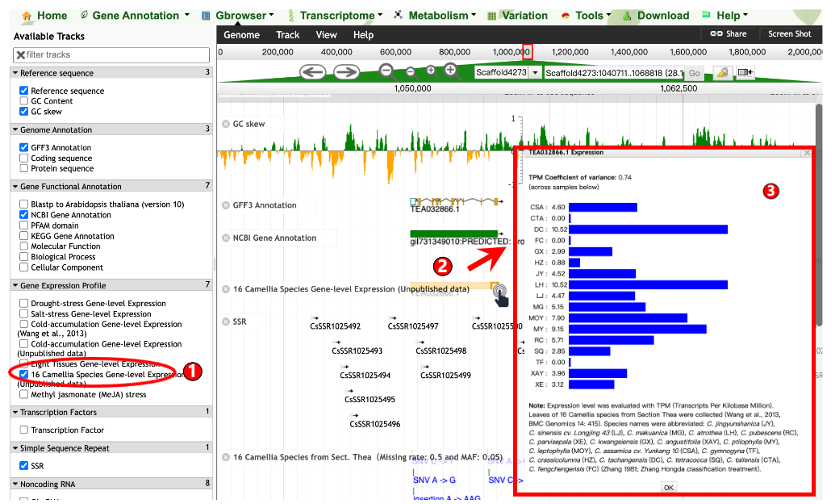
 2.1 Gbrowser
2.1 Gbrowser
The genome browser of TPIA2 provides more than 30 useful tracks to retrieve and visualize the genomic characteristics of ten Camellia genomes and their gene annotations.
 2.2 Syntenic view
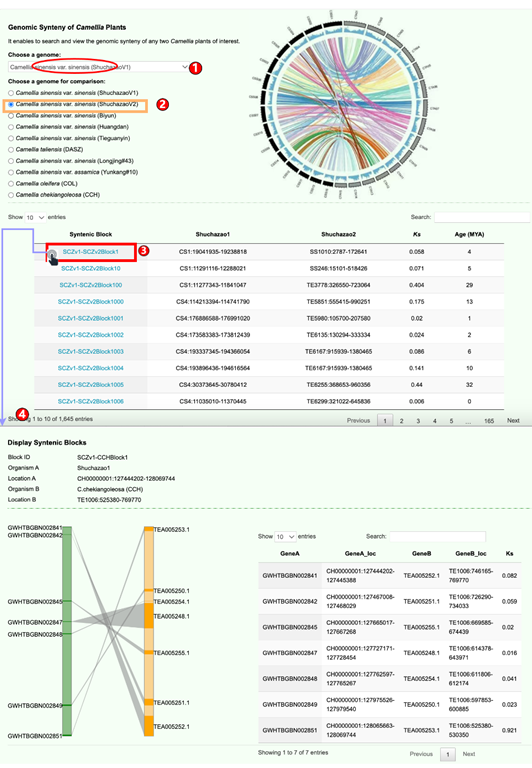
2.2 Syntenic view
TPIA2 enables users to easily search and view the genomic synteny of any two given Camellia plants of interest.
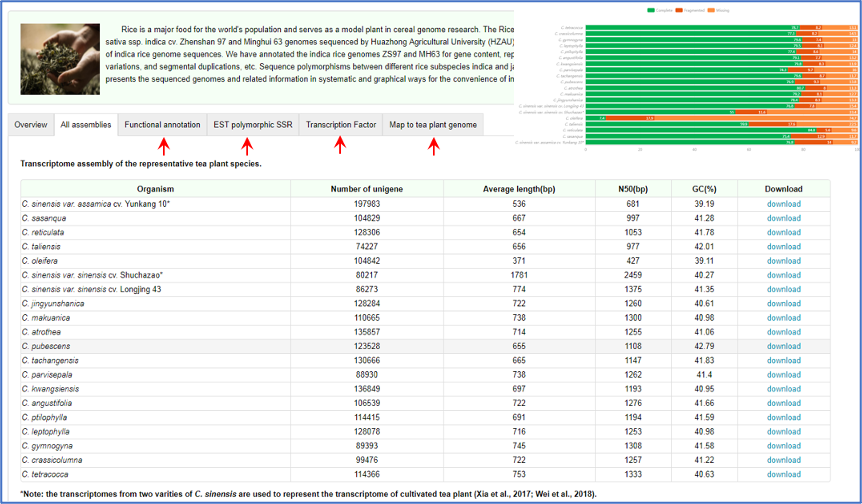
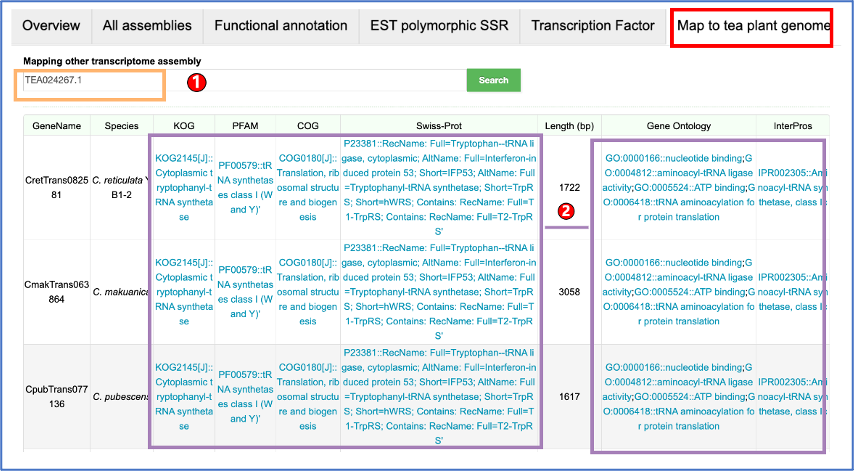
TPIA2 provides nearly all the transcriptome datasets of Camellia plants published to date (As of August 2023).

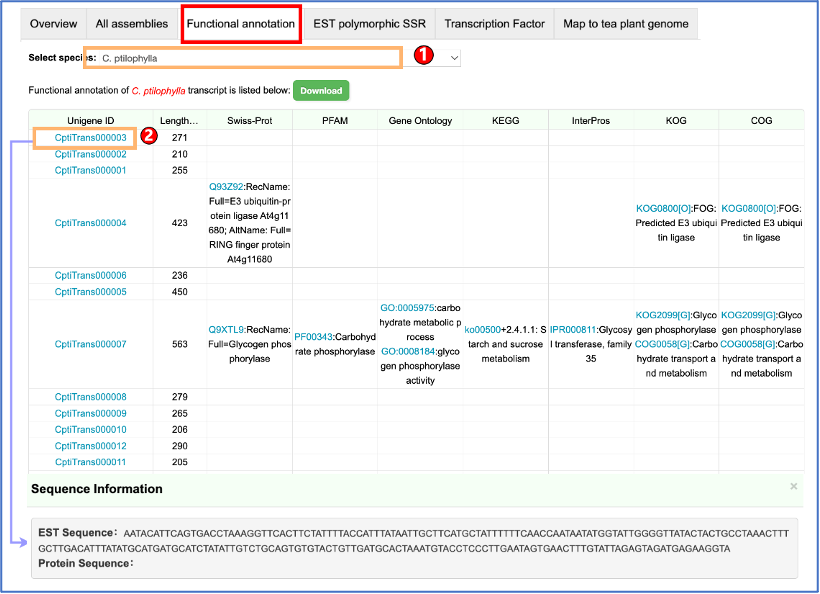
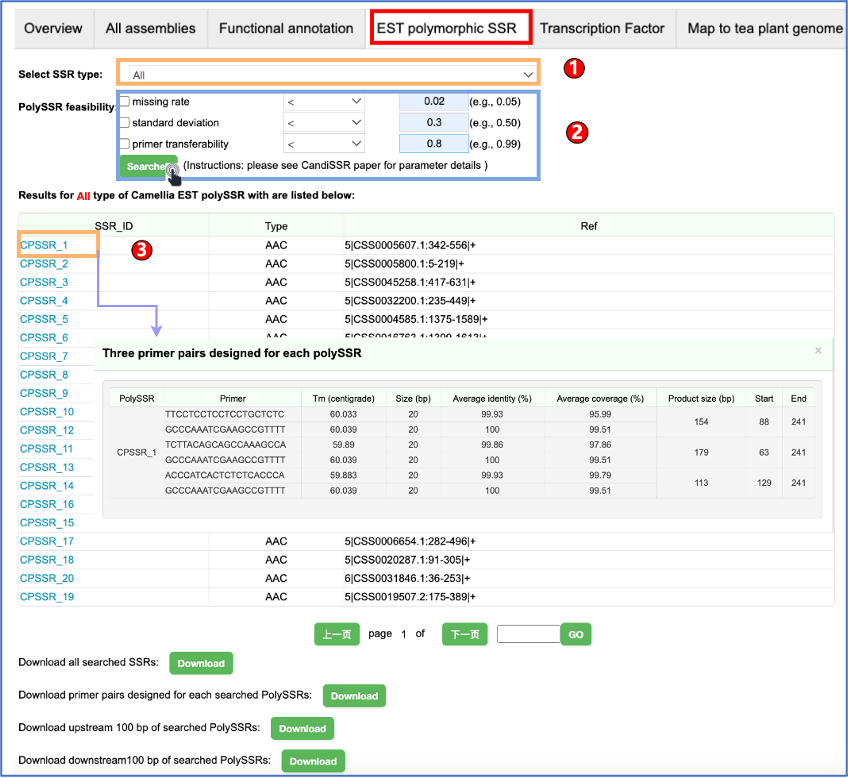
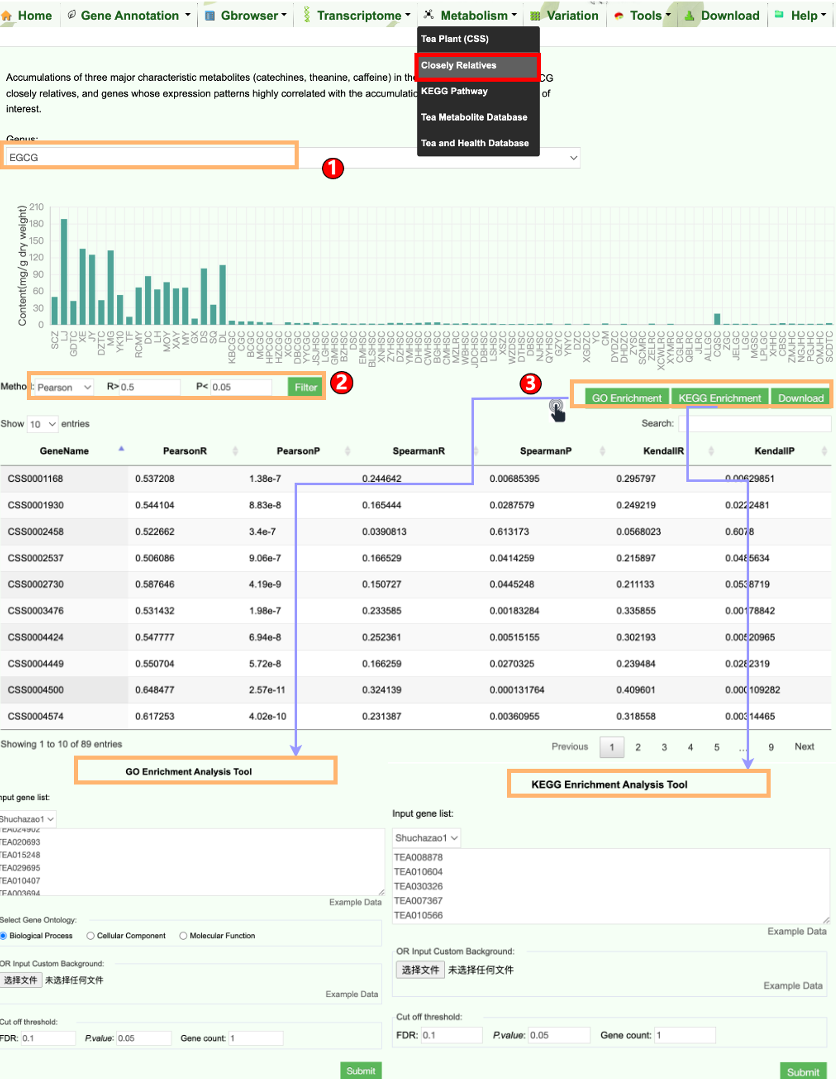
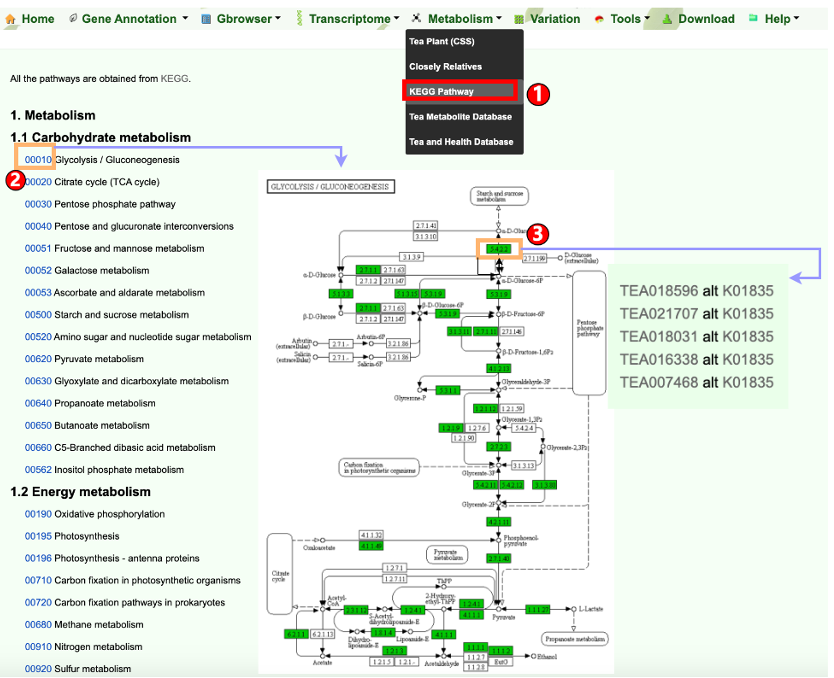
Camellia is rich in secondary metabolites that mainly include catechins, caffeine, and theanine. They not only determine the quality traits of Camellia plants, but also give Camellia with health benefits. Users can easily retrieve the metabolite data of Camellia plants as below.
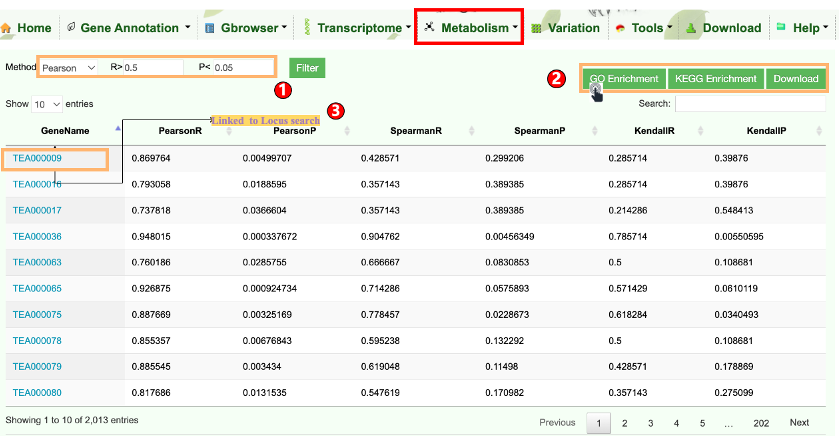
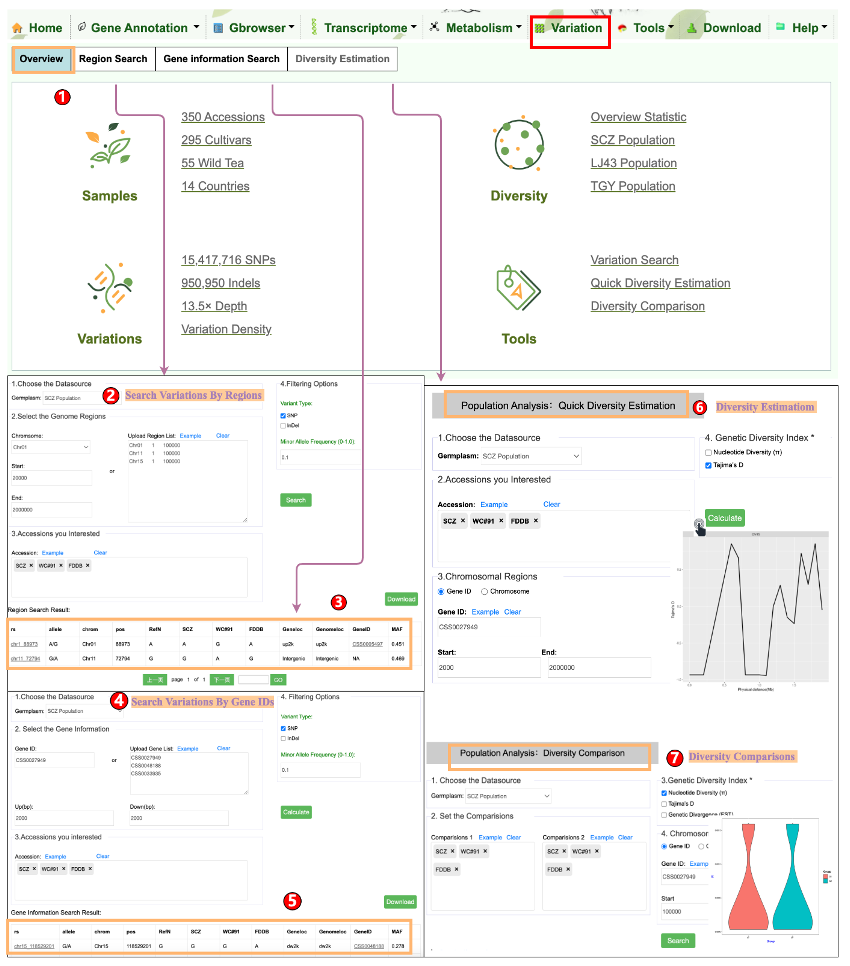
Variation module was developed for visualization of SNPs and InDels information of tea accessions. In this module, user can retrieve the genetic variation of interested genomic region(s) and gene(s) in the selected tea individuals or population.
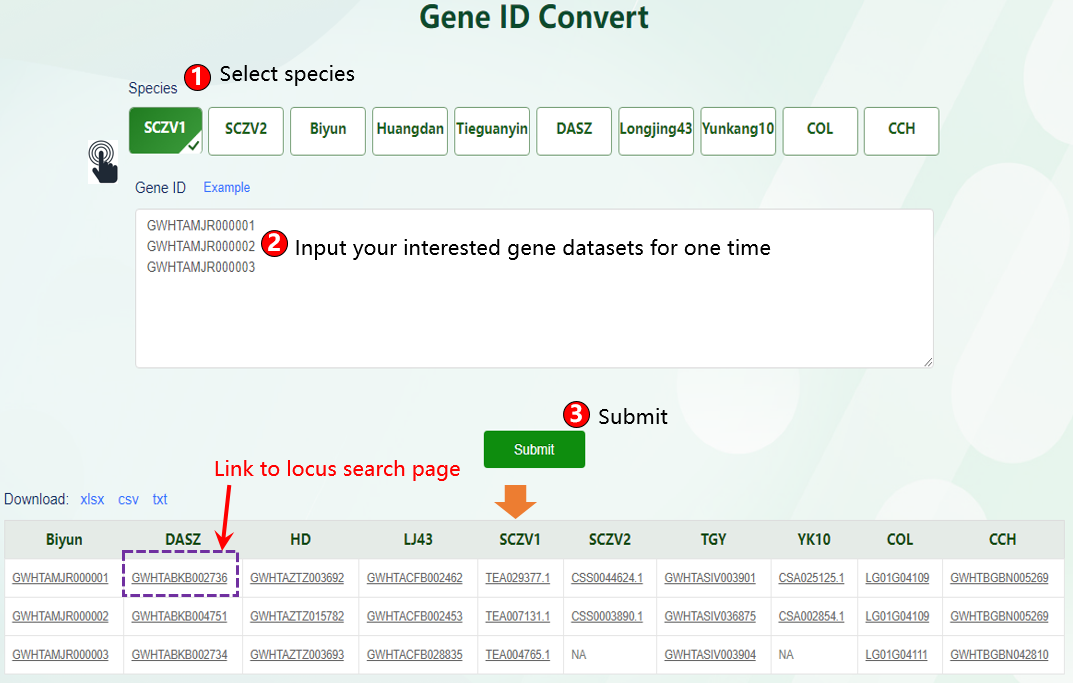
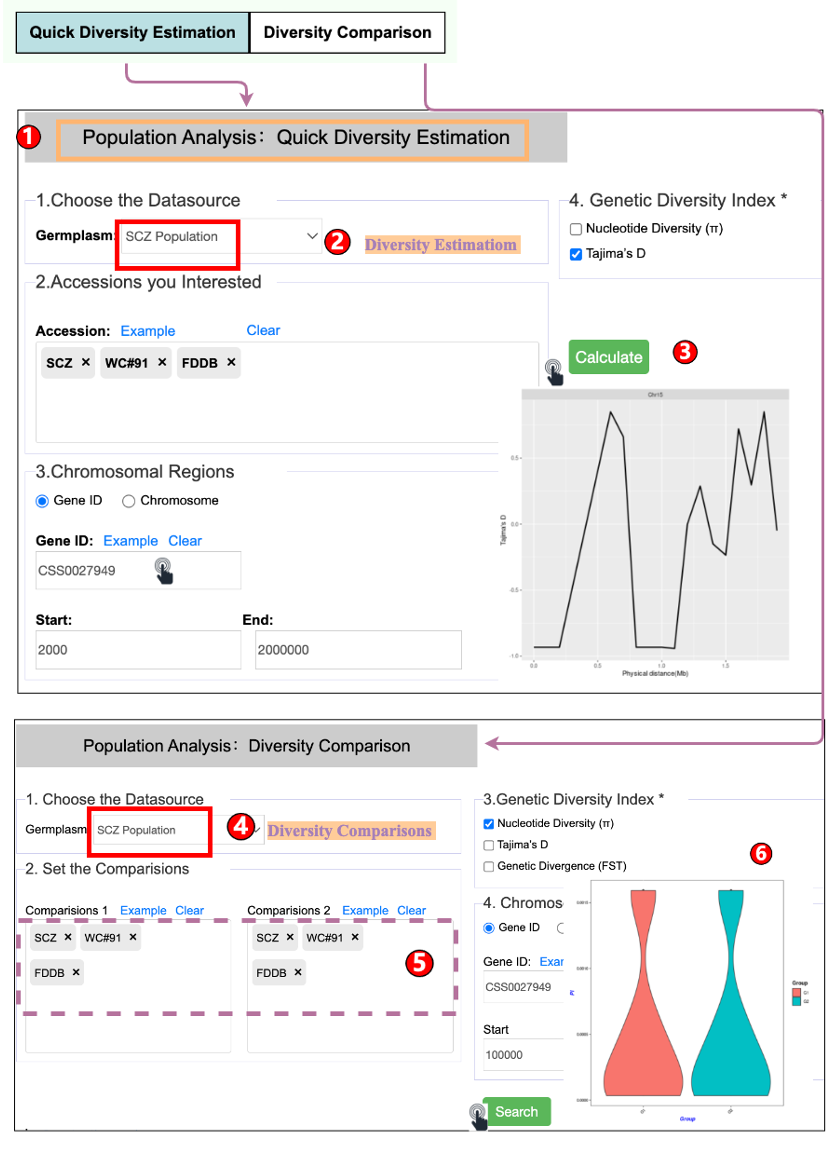
TPIA2 implements nine common but useful tools. Among them, seven are updated tools and two are newly established tools. The two newly developed tools include gene ID conversion and genetic variation analysis, which can separately facilitate rapid ID conversion of gene locus from different Camellia plants as well as data analysis of basic population genetic variants (such as π, FST, Tajima's D). The seven updated tools include KEGG and GO enrichment analysis, orthologous search, correlation analysis, ORF finder, data batch retrieval, primer design, and polySSR discovery, which are mainly updated with expanded datasets, such as with increasing expression data, orthologous, and metabolites. These newly developed or updated tools will enable users better to analyze and visualize the genomic data of tea plants and other plants, thereby promoting the genetic breeding of tea plants and other crops.
 6.1 Gene ID Convert
6.1 Gene ID Convert
 6.2 Population Genetics
6.2 Population Genetics
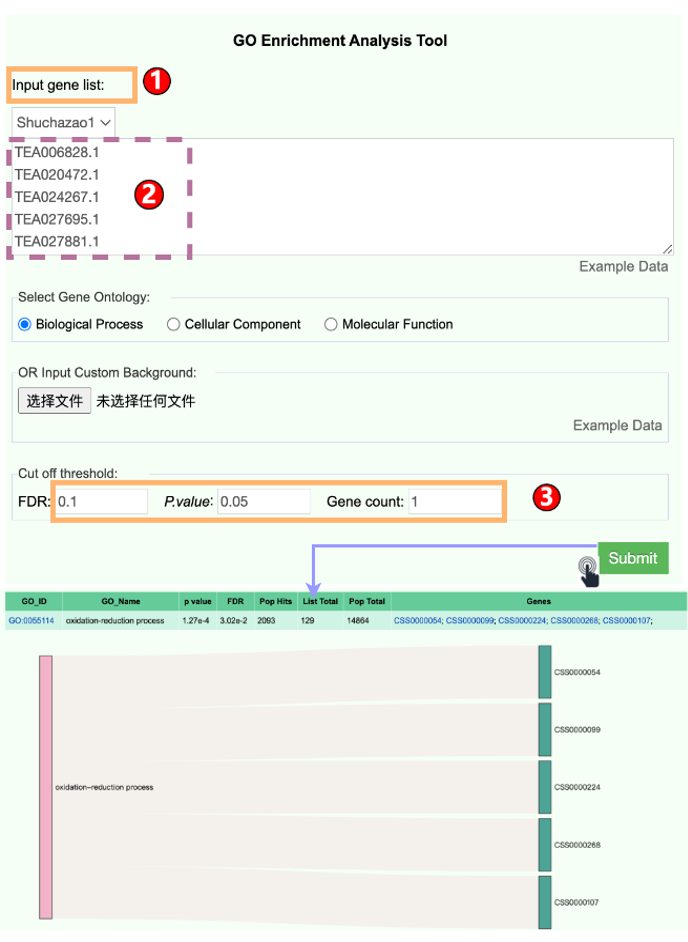
 6.3 Enrichment Analysis
6.3 Enrichment Analysis
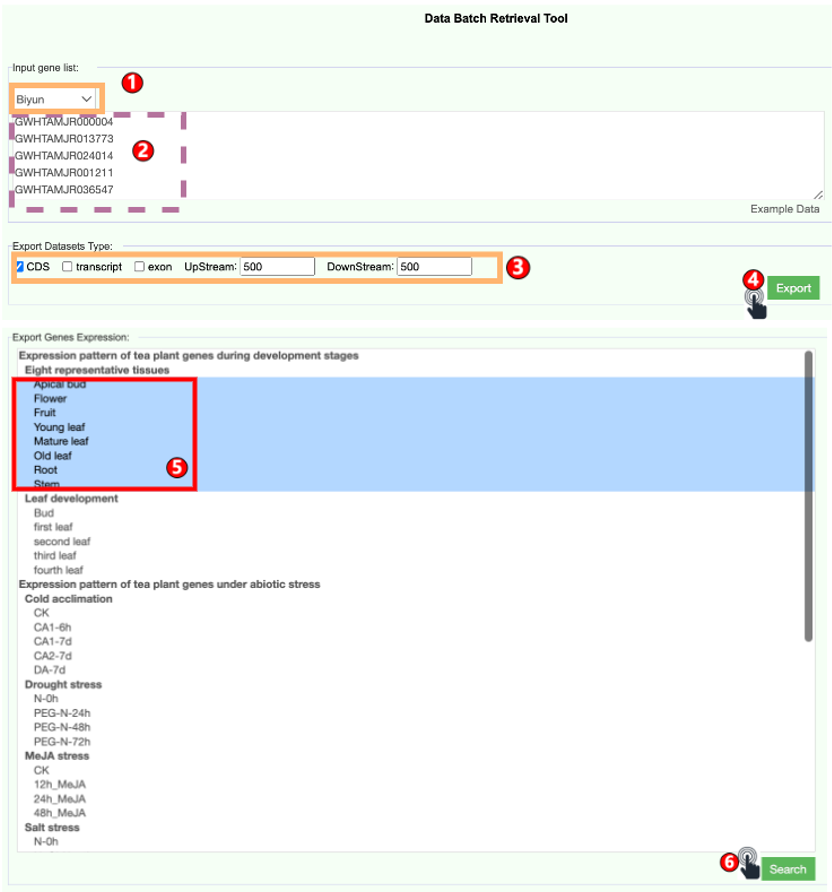
 6.4 Batch Retrieve Data
6.4 Batch Retrieve Data
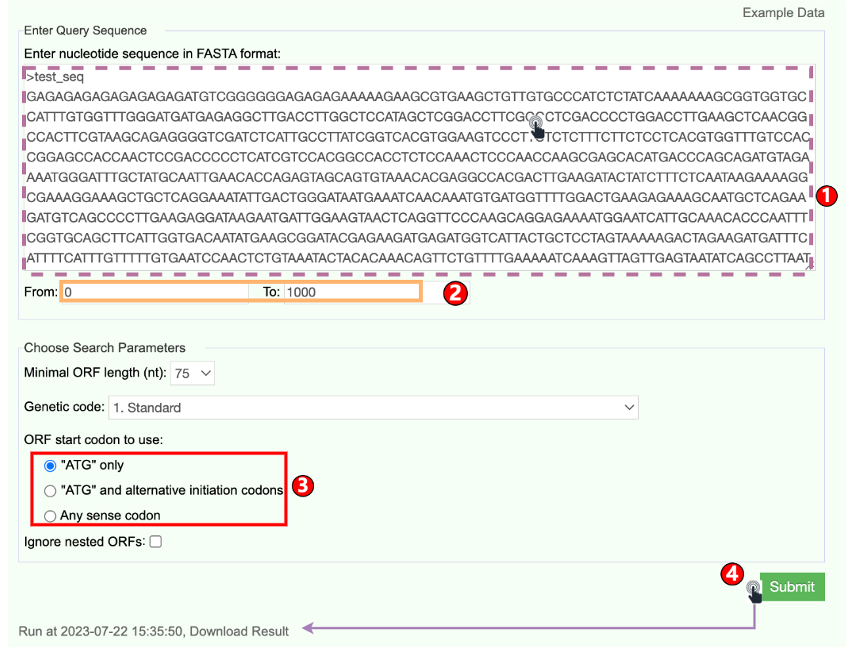
 6.5 ORF Finder
6.5 ORF Finder
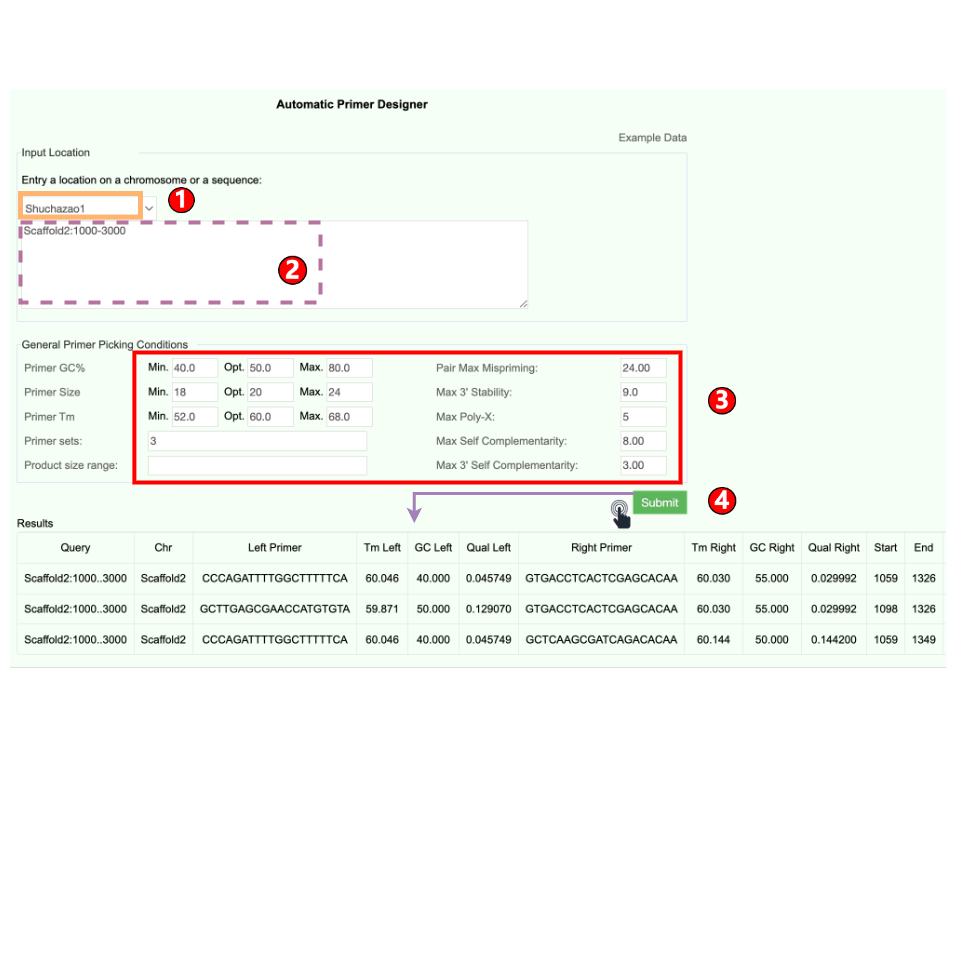
 6.6 Primer Design
6.6 Primer Design
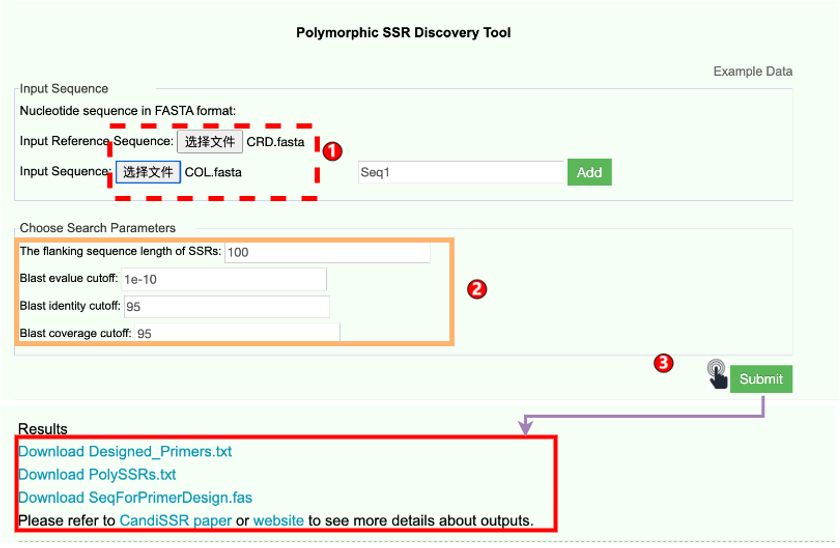
 6.7 PolySSR Design
6.7 PolySSR Design
TPIA2 is freely available to the public without any registration or login requirements (http://tpia.teaplants.cn). All the related datasets can be downloaded from the Download module of the database.
